First released in 2017, DeepVariant is an open source tool that enables researchers and clinicians to analyze an individual’s genome sequencing data and identify genetic variants, such as those that may cause disease. Our continued work on DeepVariant has been recognized for its top-of-class accuracy. With DeepTrio, we have expanded DeepVariant to be able to consider the genetic variants in the sequence data of a mother-father-child trio.
Humans are diploid organisms, carrying two copies of the human genome. Every individual inherits one copy of the genome from their mother, and the other from their father. Parental inheritance informs analysis of traits and diseases that follow Mendelian inheritance. DeepTrio learns to use the properties of Mendelian inheritance directly from sequencing data in order to more accurately identify genetic variants in cases when both parent and a child sample can be co-analyzed.
Modifying DeepVariant to analyze trio samples
DeepVariant learns to classify positions in a genome as reference or variant using representations of data similar to the “genome browser” which experts use in analysis. “Improving the Accuracy of Genomic Analysis with DeepVariant 1.0” provides a good overview.DeepVariant receives data as a window of the genome centered on a candidate variant which it is asked to classify as either reference (no variant), heterozygous (one copy of a variant) or homozygous (both copies are variant). DeepVariant sees the sequence evidence as channels representing features of the data (see: “Looking through DeepVariant’s eyes” for a deeper explanation).
We modified DeepTrio to represent the sequence data from a trio in a single image, with a fixed height for each sample and the child in the middle. Using gold standard samples from NIST Genome in a Bottle for truth labels, we train one model to call variants in the child and another to call variants in the top parent. To call both parents, we flip the position of the parent samples.
Figure 1. (top) An image of 4 of the channels that DeepTrio uses in classification (these, and 4 other channels are shown in a stack. (bottom) conceptual schematic of how trio files are used to create examples, which are then called by DeepTrio.
Measuring DeepTrio’s improved accuracy
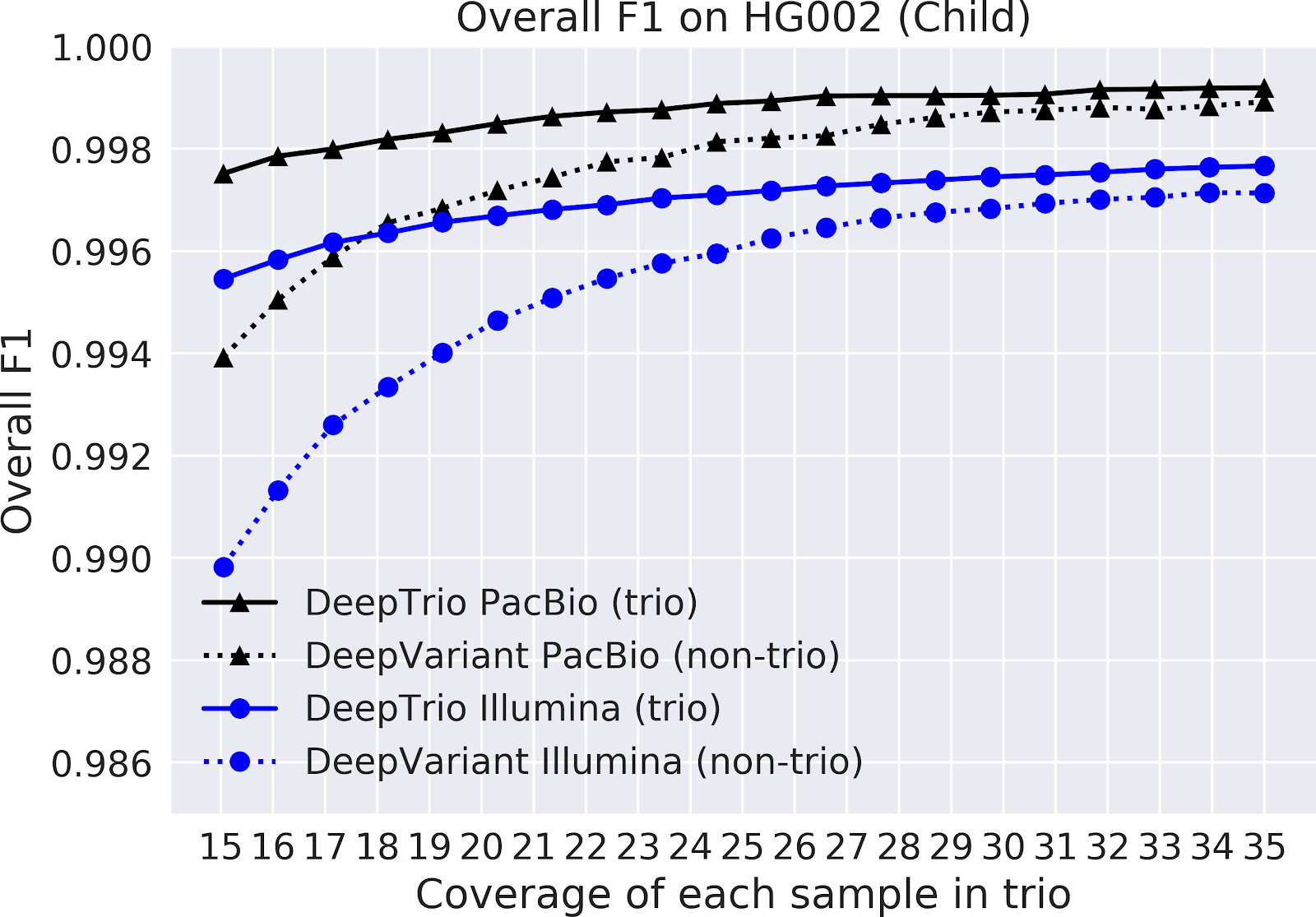
We show that DeepTrio is more accurate than DeepVariant for both parent and child variant detection, with an especially pronounced advantage at lower coverages. This enables researchers to either analyze samples at higher accuracy, or to maintain comparable accuracy at a substantially reduced expense.To assess the accuracy of DeepTrio, we compare its accuracy to DeepVariant using extensively characterized gold standards made available by NIST Genome in a Bottle. In order to have an evaluation dataset which is never seen in training, we exclude chromosome 20 from training and perform evaluations on chromosome 20.
We train DeepVariant and DeepTrio for sequencing data from two different instruments, Illumina and Pacific Biosciences (PacBio), for more information on the differences between these technologies, please see our previous blog. These sequencers both randomly sample the genome in an error-prone manner. To accurately analyze a genome, the same region needs to be sampled repeatedly. The depth of sampling at a position is called coverage. Sequencing to greater coverage is more expensive in an approximately linear manner. This often forces trade-offs between cost, accuracy, and samples sequenced. As a result, in trios parents are often sequenced at lower depth.
In the charts below, we plot the accuracy of DeepTrio and DeepVariant across a range of coverages.
Figure 2. F1-score for DeepTrio (solid line) and DeepVariant (dashed line) on a child sample (top) and a parent sample (bottom), sequenced with an Illumina (blue) and PacBio (black) instrument. F1 is measured for all types of small variants on chromosome 20, across samples with a range of sequencing coverage (x-axis).
DeepTrio’s performance on de novo variants
Each individual has roughly 5 million variants relative to the human reference genome. The overwhelming majority of these are inherited from their parents. A small number, around 100, are new (referred to as de novo), due to copying errors during DNA replication. We demonstrate that DeepTrio substantially reduces false positives for de novo variants. For Illumina data, this comes with a smaller decrease in recovery of true positives, while for PacBio data, this trade-off does not occur.To assess accuracy we analyzed sites where both parents are called as non-variant, but the child is called as heterozygous variant. We observe that DeepTrio is more reluctant to call a variant as de novo, which is similar to how a human would require a higher level of evidence for sites violating Mendelian inheritance. This results in a much lower false positive rate for these de novo variants, but a slightly lower recall rate in DeepTrio Illumina. Usually when this occurs, the child is still called as a variant, but the parents are given “no-call” (the classifier is not confident enough to make a call).
Figure 3. Accuracy on de novo calls (child heterozygous variant, parents reference call) for recall of true de novo events (top) and false positive de novo events (bottom) for DeepTrio (solid line) and DeepVariant (dashed line) on Illumina (blue) and PacBio (black). Accuracy is measured on chromosome 20, across samples with a range of sequencing coverage (x-axis).
Contributing to rare disease research
By releasing DeepTrio as open source software, we hope to improve analysis of genomic data, by allowing scientists to more accurately analyze samples. We hope this will enable research and clinical pipelines, leading to better resolution of rare disease cases, and improve development of therapeutics.In addition to the release of DeepTrio’s code as open source, we have also released the sequencing data that we generated in order to train these models. That data is described in our pre-print “An Extensive Sequence Dataset of Gold-Standard Samples for Benchmarking and Development”. By releasing both this production model, and the data required to train models of similar complexity, we hope to contribute to methods development by the genomics community.